dCBP-1
CAS No. 2484739-25-3
dCBP-1( —— )
Catalog No. M28637 CAS No. 2484739-25-3
dCBP-1 is a chemical degrader of p300/CBP. dCBP-1 hijacks the E3 ubiquitin ligase CRBN for selective degradation of p300/CBP. Degradation of p300/CBP by dCBP-1 leads to effective multiple myeloma cell killing.
Purity : >98% (HPLC)
 COA
COA
 Datasheet
Datasheet
 HNMR
HNMR
 HPLC
HPLC
 MSDS
MSDS
 Handing Instructions
Handing Instructions
| Size | Price / USD | Stock | Quantity |
| 5MG | 177 | Get Quote |


|
| 10MG | 312 | Get Quote |


|
| 25MG | 530 | Get Quote |


|
| 50MG | 758 | Get Quote |


|
| 100MG | 1044 | Get Quote |


|
| 200MG | Get Quote | Get Quote |


|
| 500MG | Get Quote | Get Quote |


|
| 1G | Get Quote | Get Quote |


|
Biological Information
-
Product NamedCBP-1
-
NoteResearch use only, not for human use.
-
Brief DescriptiondCBP-1 is a chemical degrader of p300/CBP. dCBP-1 hijacks the E3 ubiquitin ligase CRBN for selective degradation of p300/CBP. Degradation of p300/CBP by dCBP-1 leads to effective multiple myeloma cell killing.
-
DescriptiondCBP-1 is a chemical degrader of p300/CBP. dCBP-1 hijacks the E3 ubiquitin ligase CRBN for selective degradation of p300/CBP. Degradation of p300/CBP by dCBP-1 leads to effective multiple myeloma cell killing.(In Vitro):Treatment of the human haploid cell line HAP1 for 6 h with dCBP-1 revealed almost complete loss of both CBP and p300 between 10 and 1,000 nM doses. A time course analysis with 250 nM dCBP-1 revealed almost complete degradation of p300/CBP within an hour of treatment. dCBP-1 was also able to induce near-complete p300/CBP degradation across other multiple myeloma cell lines tested, including MM1R, KMS-12-BM, and KMS34.
-
In VitroWestern Blot Analysis Cell Line:Multiple myeloma cell line MM1S Concentration:10 nM, 100 nM, 250 nM, 500 nM, 1000 nM Incubation Time:6 hours Result:Revealed rapid degradation with near-complete loss of p300/CBP after 2 hours.
-
In Vivo——
-
Synonyms——
-
PathwayChromatin/Epigenetic
-
TargetEpigenetic Reader Domain
-
Recptor——
-
Research Area——
-
Indication——
Chemical Information
-
CAS Number2484739-25-3
-
Formula Weight1028.11
-
Molecular FormulaC51H63F2N11O10
-
Purity>98% (HPLC)
-
SolubilityIn Vitro:?DMSO : 50 mg/mL (48.63 mM)
-
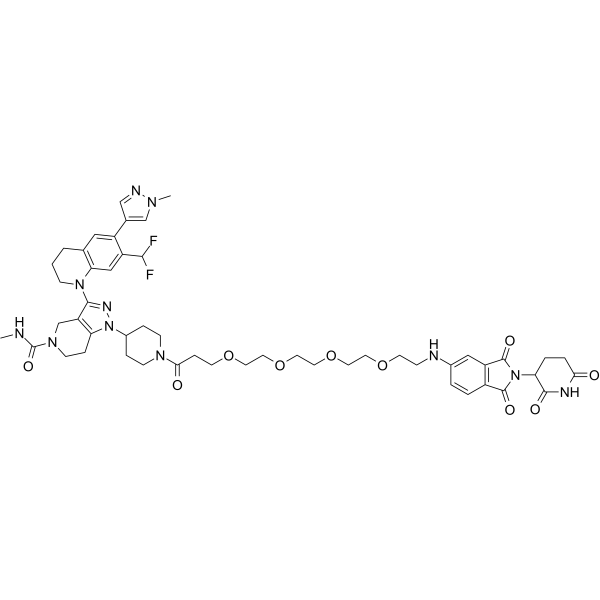
SMILESCNC(N(CC1)Cc2c1n(C(CC1)CCN1C(CCOCCOCCOCCOCCNc(cc1)cc(C(N3C(CCC(N4)=O)C4=O)=O)c1C3=O)=O)nc2N(CCCc1c2)c1cc(C(F)F)c2-c1cn(C)nc1)=O
-
Chemical Name——
Shipping & Storage Information
-
Storage(-20℃)
-
ShippingWith Ice Pack
-
Stability≥ 2 years
Reference
1.Borooah J, Leaback DH, Walker PG. Studies on glucosaminidase. 2. Substrates for N-acetyl-beta-glucosaminidase. Biochem J. 1961 Jan;78(1):106-10.
molnova catalog



related products
-
PBRM1-BD2-IN-5
PBRM1-BD2-IN-5 is a potent inhibitor of the PBRM1 Bromodomain, with dissociation constant (Kd) values of 1.5 μM and 3.9 μM for PBRM1-BD2 and PBRM1-BD5 respectively, and an inhibitory concentration 50 (IC50) value of 0.26 μM for PBRM1-BD2.
-
Molibresib besylate
Molibresib besylate (GSK 525762C) is a selective and potent inhibitor of the bromodomain and extra-terminal (BET) family of proteins with potential anticancer activity for the study of refractory hematologic malignant diseases.
-
CREB inhibitor
666-15 is a potent and selective CREB inhibitor (IC50: 81 nM).



 Cart
Cart
 sales@molnova.com
sales@molnova.com


